[1]:
#!/usr/bin/env python
Demo: trinidi.cross_section Module#
This script illustrates the functionality of the trinidi.cross_section submodule.
[2]:
from copy import deepcopy
import numpy as np
import matplotlib.pyplot as plt
from trinidi import cross_section
Available Isotopes#
First we demonstrate how to display available isotopes and display their energy ranges.
The cross_section.info function prints out info for all isotopes.
[3]:
# cross_section.info() # Uncomment this line to print the complete output.
# H-1 10.0 µeV to 20.0 MeV
# H-2 10.0 µeV to 150.0 MeV
# H-3 10.0 µeV to 20.0 MeV
# He-3 10.0 µeV to 20.0 MeV
# He-4 10.0 µeV to 20.0 MeV
# Li-6 10.0 µeV to 20.0 MeV
# Li-7 10.0 µeV to 20.0 MeV
# Be-7 10.0 µeV to 20.0 MeV
# Be-9 10.0 µeV to 20.0 MeV
# B-11 10.0 µeV to 20.0 MeV
# C-12 10.0 µeV to 150.0 MeV
# .
# .
# .
We access a list of all available isotopes using the cross_section.avail function.
[4]:
av_isotopes = cross_section.avail()
print(
f"av_isotopes = [{av_isotopes[0]}, {av_isotopes[1]}, ..., {av_isotopes[-2]}, {av_isotopes[-1]}]"
)
print(f"Number of isotopes = {len(av_isotopes)}")
av_isotopes = [H-1, H-2, ..., Es-255, Fm-255]
Number of isotopes = 533
You can restrict the displayed isotopes by cross_section.info using the isotopes optional argument. Below we subselect all the uranium isotopes.
[5]:
isotopes_U = [iso for iso in av_isotopes if iso.split("-")[0] == "U"]
print("\nUranium Isotopes:")
cross_section.info(isotopes=isotopes_U)
Uranium Isotopes:
U-230 10.0 µeV to 20.0 MeV
U-231 10.0 µeV to 20.0 MeV
U-232 10.0 µeV to 20.0 MeV
U-233 10.0 µeV to 20.0 MeV
U-234 10.0 µeV to 30.0 MeV
U-235 10.0 µeV to 30.0 MeV
U-236 10.0 µeV to 30.0 MeV
U-237 10.0 µeV to 30.0 MeV
U-238 10.0 µeV to 30.0 MeV
U-239 10.0 µeV to 30.0 MeV
U-240 10.0 µeV to 30.0 MeV
U-241 10.0 µeV to 30.0 MeV
Generate and Plot a Cross Section Dictionary Object XSDict#
First we setup the time-of-flight (TOF) array and requested isotopes list. Note that the cross section dictionary is sampled with equispaced samples in TOF.
[6]:
Δt = 0.30 # bin width [μs]
flight_path_length = 10 # [m]
t_F = np.arange(72, 720, Δt) # time-of-flight array [μs]
isotopes = ["U-235", "U-238"]
Create a XSDict cross section dictionary object.
[7]:
D = cross_section.XSDict(isotopes, t_F, flight_path_length)
print(D)
<class 'trinidi.cross_section.XSDict'>
isotopes = ['U-235', 'U-238']
N_m = 2
t_F = [72.000 μs, ..., 719.700 μs]
Δt = 0.300 μs
N_F = 2160
flight_path_length = 10.000 m
E = [100.830 eV, ..., 1.009 eV]
samples_per_bin = 10
values.shape = (2, 2160) = (N_m, N_F)
The np.ndarray D.values is a matrix of size N_m x N_F that contains the cross section values.
[8]:
print(f"{D.values.shape = }")
D.values.shape = (2, 2160)
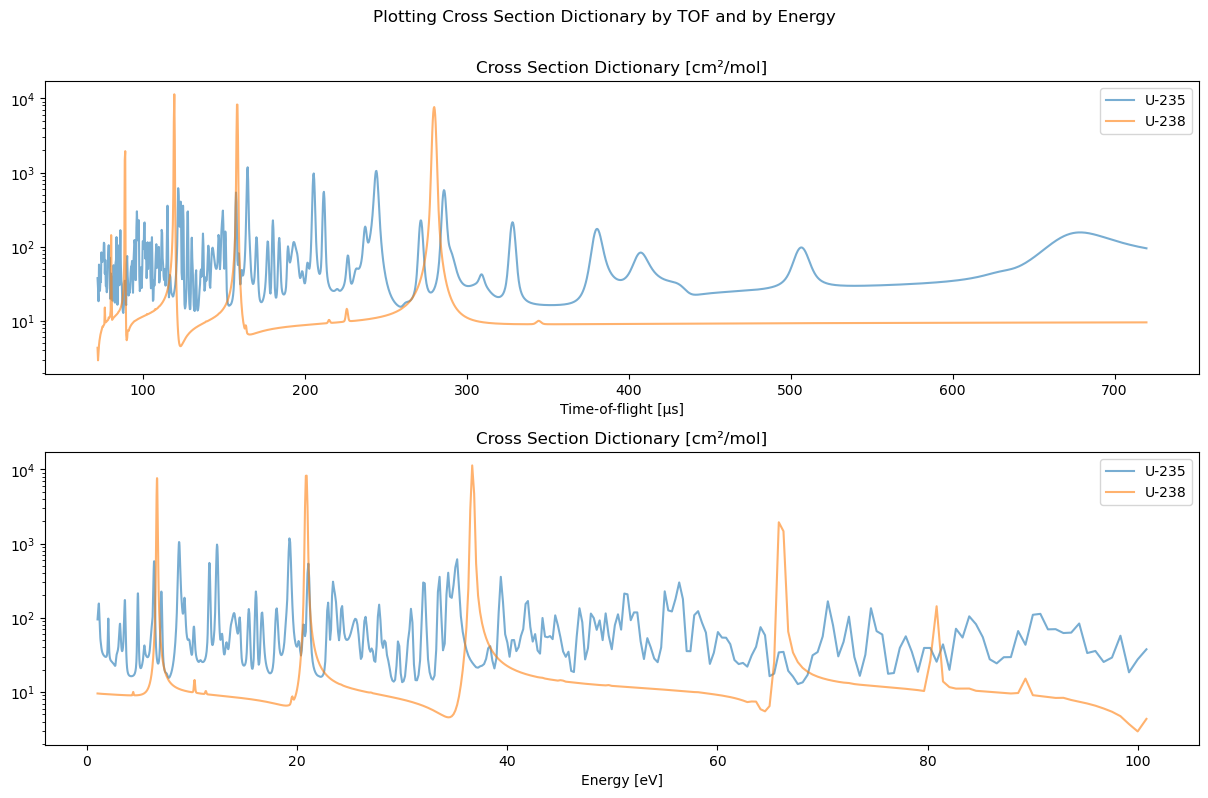
You can plot the dictionary easily with the XSDict.plot function. The optional argument function_of_energy=False [default] allows plotting it as a function of time-of-flight while function_of_energy=True plots it as a function of energy.
[9]:
fig, ax = plt.subplots(2, 1, figsize=[12, 8], sharex=False)
ax = np.atleast_1d(ax)
D.plot(ax[0])
D.plot(ax[1], function_of_energy=True)
fig.suptitle("Plotting Cross Section Dictionary by TOF and by Energy")
plt.tight_layout(pad=0.4, w_pad=0.5, h_pad=1.0, rect=(0, 0, 1, 0.95))
plt.show()
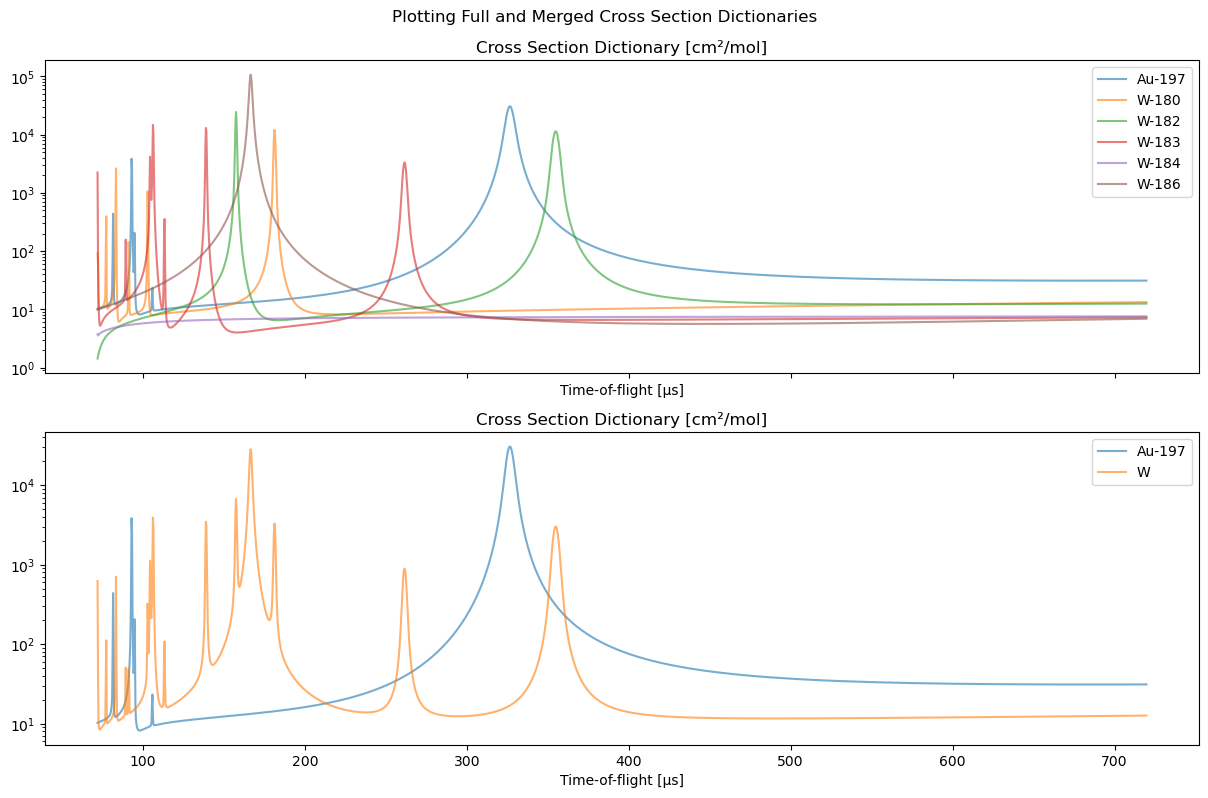
Creating a Compound Material Cross Section Dictionary#
When reconstructing a sample with isotopes in known proportions, we recommend combining the corresponding isotopes according to their known abundances and treating it as a compound material.
For example, below we show how we construct a gold and tungsten (Au, W) cross section dictionary. Gold is practically 100% Au-197 so no action is necessary. However, elemental tungsten consists of several isotopes,
[10]:
# W-180: 0.12%
# W-182: 26.5%
# W-183: 14.3%
# W-184: 30.6%
# W-186: 28.4%
which we want to combine into a single entry with the use of the XSDict.merge function.
We start out with the XSDict using all isotopes. The D_full object will stay untouched and used for later comparison.
[11]:
isotopes_full = ["Au-197", "W-180", "W-182", "W-183", "W-184", "W-186"]
D_full = cross_section.XSDict(isotopes_full, t_F, flight_path_length)
We now define the arguments for the XSDict.merge function. The arguments below result in a new cross section that will be
and a new dictionary that will be
[12]:
merge_isotopes = ["W-180", "W-182", "W-183", "W-184", "W-186"]
merge_weights = [0.0012, 0.265, 0.143, 0.306, 0.284]
new_key = "W"
Below, we generate the modified XSDict object, D_new from a copy of D_full.
[13]:
D_new = deepcopy(D_full)
isotopes_new = D_new.merge(merge_isotopes, merge_weights, new_key)
print(f"New isotope list: {isotopes_new}")
print(D_new)
New isotope list: ['Au-197', 'W']
<class 'trinidi.cross_section.XSDict'>
isotopes = ['Au-197', 'W']
N_m = 2
t_F = [72.000 μs, ..., 719.700 μs]
Δt = 0.300 μs
N_F = 2160
flight_path_length = 10.000 m
E = [100.830 eV, ..., 1.009 eV]
samples_per_bin = 10
values.shape = (2, 2160) = (N_m, N_F)
The merge_isotopes cross sections are combined using a weighted sum using the merge_weights. All previous isotope keys, ["W-180", "W-182", "W-183", "W-184", "W-186"], will be replaced by the new_key, "W", which is defined by the user.
(Note that the updated list XSDict.isotopes is now not necessarily strictly isotopes since "W" is not an isotope. After creation this list primarily serves for plotting and identification of the entries.)
Below we plot and compare the resulting XSDict objects.
[14]:
fig, ax = plt.subplots(2, 1, figsize=[12, 8], sharex=True)
ax = np.atleast_1d(ax)
D_full.plot(ax[0])
D_new.plot(ax[1])
plt.tight_layout(pad=0.4, w_pad=0.5, h_pad=1.0, rect=(0, 0, 1, 0.95))
fig.suptitle("Plotting Full and Merged Cross Section Dictionaries")
plt.show()